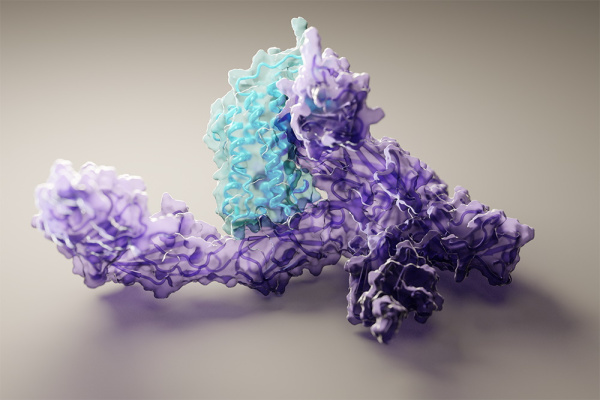
Now researchers claim to have leapfrogged DeepMind the way DeepMind leapfrogged the rest of the world, with RoseTTAFold, a system that does nearly the very same thing at a fraction of the computational cost. Examples of anticipated protein structures and their ground realities.” We have a public server that anyone can send protein sequences to and have the structures predicted,” Baker said. You may recall Folding@Home, the popular distributed computing app that let individuals donate their computing cycles to trying to predict protein structures. We are already utilizing RoseTTAFold for protein design and more systematic protein-protein complex structure prediction, and we are excited about rapidly improving these, along with standard single chain modeling, by integrating ideas from the DeepMind paper.
That issue appears to have actually been at least partly mooted by work from University of Washington researchers led by David Baker and Minkyung Baek, published in the most recent concern of the journal Science. Baker, you may remember, recently won a Breakthrough Prize for his teams work combating COVID-19 with crafted proteins.
The teams brand-new design, RoseTTAFold, makes predictions at comparable precision levels using approaches that Baker, responding to concerns through e-mail, openly confessed were influenced by those utilized by AlphaFold2.
” The AlphaFold2 group presented numerous new top-level principles at the CASP14 conference. Starting from these concepts, and with a lot of collective brainstorming with colleagues in the group, Minkyung has been able to make fantastic progress in very little time,” he stated.
Examples of anticipated protein structures and their ground realities. A rating above 90 is thought about exceptionally great. Image Credits: UW/Baek et al
. Bakers group basically placed 2nd at CASP14, no mean task, however hearing DeepMinds approaches described even generally set them on a collision course. They established a “three-track” neural network that at the same time thought about the amino acid sequence (one measurement), distances in between residues (two dimensions) and collaborates in space (three measurements). The application is beyond complex and far outside the scope of this article, however the result is a model that accomplishes almost the very same accuracy levels– levels, it bears duplicating, that were totally extraordinary less than a year ago.
Whats more, RoseTTAFold accomplishes this level of precision even more rapidly– that is, using less computation power. As the paper puts it:.
DeepMind reported using several GPUs for days to make specific forecasts, whereas our predictions are made in a single pass through the network in the exact same way that would be utilized for a server … the end-to-end version of RoseTTAFold requires ~ 10 minutes on an RTX2080 GPU to create backbone collaborates for proteins with less than 400 residues.
Hear that? Its the sound of countless microbiologists sighing in relief and disposing of drafts of e-mails requesting supercomputer time. It may not be simple to lay ones hands on a 2080 nowadays, but the point is any high-end desktop GPU can perform this job in minutes, instead of requiring a high-end cluster running for days.
The modest requirements make RoseTTAFold appropriate for public hosting and circulation as well, something that may never have been in the cards for AlphaFold2.
” We have a public server that anybody can submit protein series to and have the structures predicted,” Baker said. “There have actually been over 4,500 submissions because we put the server up a few weeks ago. We have also made the source code freely available.”.
This might seem very specific niche, and it is, however protein folding has traditionally been among the toughest problems in biology and one towards which numerous hours of high-performance computing have been dedicated. You might remember Folding@Home, the popular dispersed computing app that let people donate their computing cycles to attempting to predict protein structures. The sort of issue that might have taken a thousand computer systems days or weeks to do– basically by brute-forcing options and inspecting for fit– now can be performed in minutes on a single desktop.
The physical structure of proteins is of utmost importance in biology, as it is proteins that do the large majority of tasks in our bodies, and proteins that need to be modified, suppressed, boosted and so on for therapeutic factors; initially, nevertheless, they need to be understood, and until November that understanding might not be dependably accomplished computationally. At CASP14 it was proven to be possible, and now it has been made widely readily available.
It is not, by a long shot, a “service” to the issue of protein folding, though the sentiment has actually been expressed. They contort and twist to grab or release other molecules, to block or slip through gates and other proteins, and normally to do everything they do.
” There are lots of amazing chapters ahead … the story is simply starting,” stated Baker.
Regarding the DeepMind paper, Baker provided the list below remark in the spirit of college sociability:.
Ive gone through, and think this is a stunning paper explaining fantastic work.
The DeepMind paper is really complementary to our paper, and I think it is proper that it is not coming out after ours, as our work is actually based upon their advances.
I believe that readers will take pleasure in reading both documents– they are really far from being duplicative. As we mention in our paper, their technique is more accurate than ours, and now it will be extremely intriguing to see what features of their technique are accountable for the staying differences. We are already utilizing RoseTTAFold for protein design and more systematic protein-protein complex structure prediction, and we are excited about quickly enhancing these, in addition to standard single chain modeling, by incorporating ideas from the DeepMind paper.
If youre curious about the science and the prospective consequences, think about reading this a lot more technical and in-depth account of the approaches and possible next steps composed in the wake of AlphaFold2s CASP14 performance.
DeepMind stunned the biology world late in 2015 when its AlphaFold2 AI model anticipated the structure of proteins (a typical and extremely hard problem) so precisely that numerous declared the decades-old problem “fixed.” Now scientists declare to have actually leapfrogged DeepMind the way DeepMind leapfrogged the rest of the world, with RoseTTAFold, a system that does almost the same thing at a fraction of the computational expense. (Oh, and its complimentary to use.).
AlphaFold2 has been the talk of the market given that November, when it blew away the competitors at CASP14, a virtual competition in between algorithms built to forecast the physical structure of a protein given the series of amino acids that make it up. The model from DeepMind was so far ahead of the others, so extremely and dependably accurate, that lots of in the field have actually talked (half-seriously and in great humor) about moving on to a brand-new field.
However one aspect that seemed to satisfy no one was DeepMinds plans for the system. It was not exhaustively and freely explained, and some anxious that the company (which is owned by Alphabet/Google) was intending on more or less keeping the secret sauce to themselves– which would be their authority but likewise somewhat versus the principles of mutual aid in the clinical world.
Update: In something of a surprise, DeepMind published more detailed techniques in the journal Nature today. Ive likewise included a comment from that group at the bottom of the article.